CellRank 2: Unified fate mapping in multiview single-cell data#
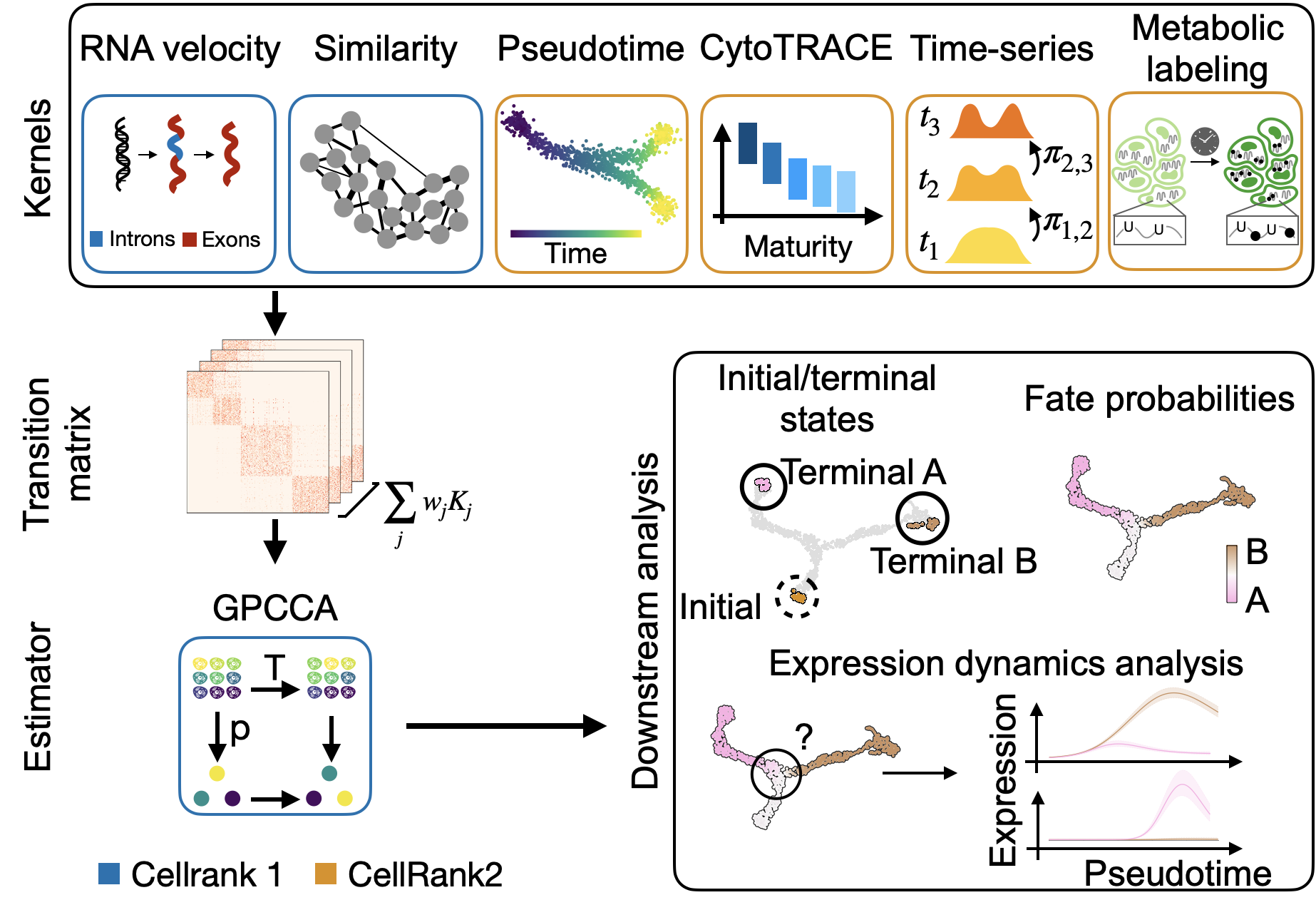
CellRank [Lange et al., 2022, Weiler et al., 2023] is a modular framework to study cellular dynamics based on Markov state modeling of multi-view single-cell data. See about CellRank to learn more and our citation guide for guidance on citing our work correctly. Also, read our recent preprint to see the new CellRank 2 features in action.
CellRank scales to large cell numbers, is fully compatible with the scverse ecosystem, and is easy to use. In the backend, it is powered by the pyGPCCA package [Reuter et al., 2019, Reuter et al., 2022]. Feel free to open an issue or send us an email if you encounter a bug, need our help or just want to make a comment/suggestion.
Important
If you’re moving from CellRank 1 to CellRank 2, check out Moving to CellRank 2.
CellRank’s Key Applications#
Estimate differentiation direction based on a varied number of biological priors, including pseudotime, developmental potential, RNA velocity, experimental time points, and
more.Compute initial, terminal and intermediate macrostates [Reuter et al., 2019, Reuter et al., 2022].
Visualize and cluster gene expression trends.
… and much more, check out our API.
Getting Started with CellRank#
We have Tutorials to help you getting started. To see CellRank in action, explore our manuscript [Lange et al., 2022] in Nature Methods.
Contributing#
We actively encourage any contribution! To get started, please check out the Contributing Guide.




